This website uses cookies to ensure a better user experience.
To get more information, please read our Cookie Statement.
Discrimination of DNA−Protein Complexes by Translocation Through Nanocapillaries
Team leader prof. A. Rađenović from EPFL, Switzerland and Sanjin Marion, assistant from Institut za fiziku, Zagreb, together with other coworkers have published a paper in Nano Letters on translocation of macromolecules through nano-sized pores. This work is a proof of concept for discriminating between different DNA-protein complexes and simultaneously pin-pointing their binding sites – a step towards practical nanopore devices.
Single Molecule Localization and Discrimination of DNA-Protein Complexes by Controlled Translocation Through Nanocapillaries
Roman D. Bulushev*, Sanjin Marion*, Ekaterina Petrova*, Sebastian J. Davis*, Sebastian J. Maerkl, Aleksandra Rađenović
Nano Letters (2016)
doi: 10.1021/acs.nanolett.6b04165
DNA-protein interactions regulate almost every aspect of cellular function, including DNA replication, transcription and maintenance: proteins search a specific DNA target, bind to it and are then able to perform a variety of functions crucial for life. In order to understand these functions on a molecular level, it is important to understand the details of how various proteins bind to DNA. This latest paper is a result of an ongoing collaboration of EPFL and IF started within the UKF project Confined DNA. Our S. Marion contributed to the paper by providing the theoretical background necessary for interpreting the experimental results. This newest work presents a promising and practical method to identify DNA-protein complexes as well as accurately find their binding sites in physiological conditions.
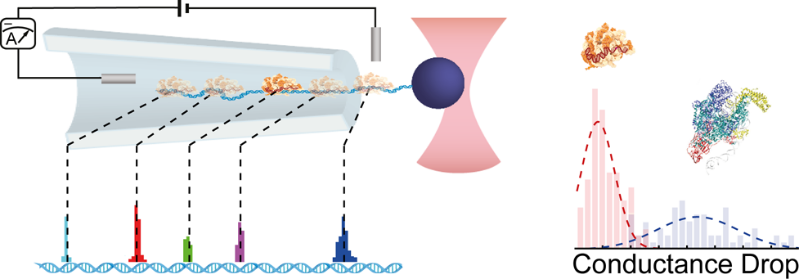
By using optical tweezers the team was able to perform controlled translocations of proteins bound on single DNA molecule through glass nanocapillaries. Two different proteins were used: RNA polymerase which is critical for the initiation of DNA-to-RNA transcription, as well as dCas9, a protein which can be programmed to bind at different DNA sites and is part of the CRISPR-Cas9 complex famous for its use in DNA editing. The programmability of dCas9 binding was used to arrange DNA binding sites as an actual ruler for benchmarking the accuracy of localization. Measuring and modeling force versus dislocation curves uncovered the non-equilibrium work done by the translocation of the protein, which in turn provided an estimate of the effective charge of the complexes using the Jarzynski equality. Thanks to its relatively robust implementation and tunability of nanocapillaries, this technique provides a new analytical tool to study protein-DNA interactions on a single molecule level.