This website uses cookies to ensure a better user experience.
To get more information, please read our Cookie Statement.
Shapes of DNA ropes condensed within viruses
A new paper by Antonio Šiber published in Scientific Reports models the structure and geometry of DNA molecules within viruses. The new work comprises not only the known shapes of packed DNA but also predicts certain novel conformations, which helps explain the functioning of viruses.
Shapes of minimal-energy DNA ropes condensed in confinement
Antonio Šiber
Scientific Reports 6, 29012 (2016).
doi: 10.1038/srep29012
Today, the shape of a virus protein capsid can experimentally be determined down to the atomic resolution of about 0.3 nm. However, less is known about the structure and geometry of DNA molecules within viruses. This is partially due to packed DNA probably having a different symmetry than the icosahedral protein capsid, so the assumption of icosahedral symmetry while averaging the experimental structural data leads to loss of information about the DNA. With this in mind, theoretical calculations provide a much-needed insight into the structure of packed DNA as well as further our understanding of viruses in general.
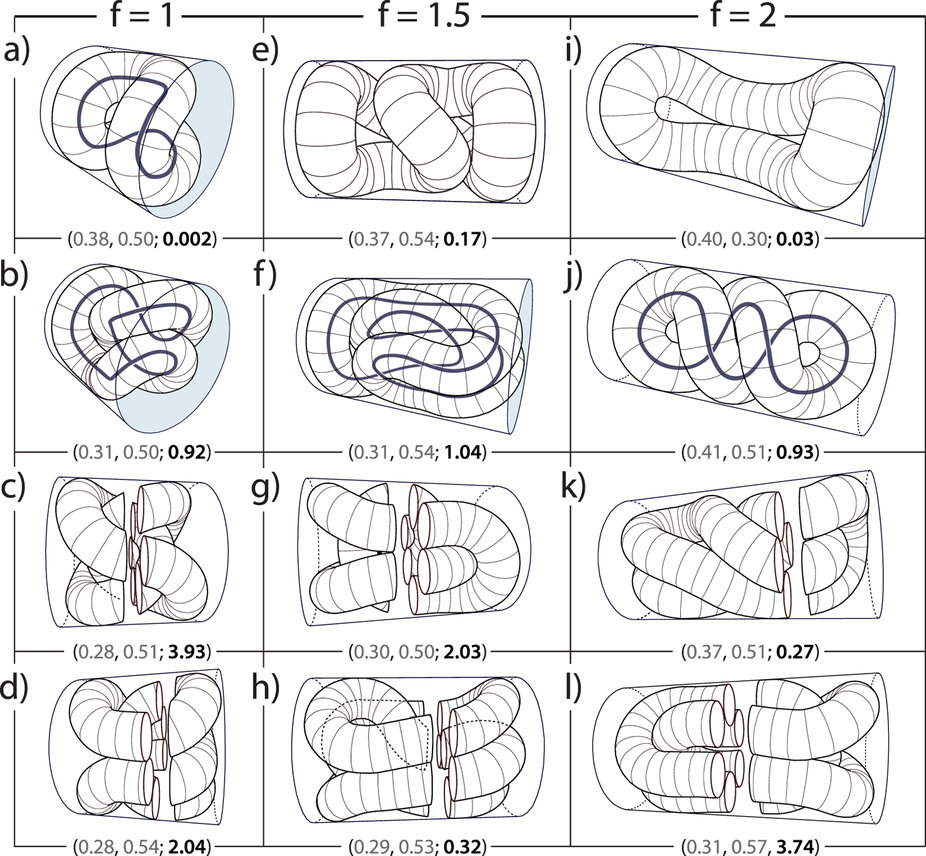
Our colleague Antonio Šiber has recently published a paper in Scientific Reports where he describes long molecules of DNA packed within a virus. The DNA is modeled as rope-like bundles which condense due to self-attracting interactions. The shape of these DNA ropes optimizes the free energy which accounts for the interplay of the bending energy and the surface energy of ropes. The writhing shapes predicted by the model depend on the shape of the confinement, the total amount of the packed DNA, but also on the relative contributions of the bending and surface energies. Many previously known conformations were successfully recovered by the model. Even though some solutions were previously thought to be contradictory, the results show they are physically viable after all and that there are many possible packing conformations of the DNA: the one which realizes in a particular virus depends on the capsid geometry and the nature of condensing agents.
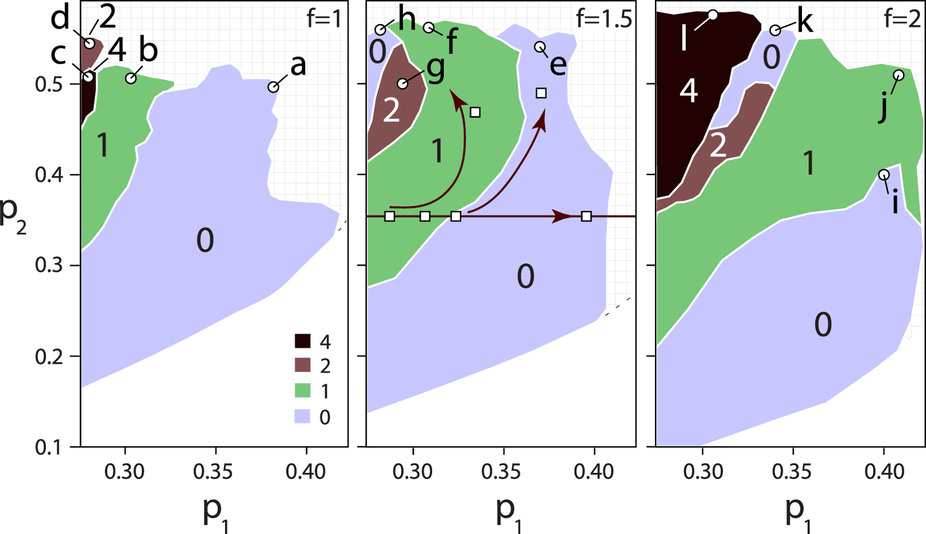
DNA rope writhe (a measure of its topological complexity) as a function of width (p1) and volume (p2) of the rope which are normalized to the protein capside width and volume, respectively.